Survival Analysis 2: Non-Parametric Estimation of Survival Functions
Concepts of survival function estimations and corresponding calculations both manually and in R.
Introduction
In the Survival Analysis 1: Basic concepts and three fundamental functions I
shared about the three basic blocks of survival analysis: density function,
survival function and hazard function.
Next, it is natural to think about: now we know their meaning, but how
to estimate them? It brings us to the next topic of non-parametric estimation
of survival functions. The word non-parametric means there is no
assumption about the distribution of survival time T.
A simple case without censoring
No censoring indicates all samples are observed until they experience the event of
interest (e.g., death, unemployment, account deletion). Then we can
estimate survival function S(t) by:
Assume we are observing 8 mice’ survival time
| Time | Accumulate death number | |
|---|---|---|
| 2 | 3 | |
| 4 | 5 | |
| 12 | 8 | = 0 |
Estimation with consoring
The method above is applicable only in situations where all individuals
are observed until the events of interest occur (no censoring). However,
in real life, such a scenario is rare because of unfeasible study
design, high expenses, etc. As a result, we have to adjust the risk
set accordingly when new observations are entering the study or
previous ones are being censored. Two common methods are Life-Table
(Actuarial) and Product-Limit (Kaplan-Meier) methods.
The Life-Table (Actuarial) method
This method objectively defines the time intervals by split points and
assume the observations being censored within an interval were at risk
for half of the length of that interval. Let me first introduce some
jargon:
- The split points =
- Then the time intervals =
- The number of observations enter the interval =
- The number of observations experiencing the event in the
interval = - The number of observations censored in the interval =
- Therefore we have
- The risk set (observations exposed to risk of experiencing the event)
= . The “” part comes from the
assumption mentioned above.
Next, we can compute conditional probability of experiencing the
event in the interval:
The conditional probability of surviving the interval:
Finally we can compute the survivor function as
- when r = 0:
- for r = 1, 2, 3, …
- with standard error estimation
- density function
- with standard error estimation
- the hazard function is estimated by
- with standard error estimation
A simple exercise
The following table contains data on the recidivism-example in Allison
(2010, p. 45):
| interval_start | num_entering | num_withdrawn | num_exposed_risk | num_events |
|---|---|---|---|---|
| 0 | 432 | 0 | 432 | 14 |
| 10 | 418 | 0 | 418 | 21 |
| 20 | 397 | 0 | 397 | 23 |
| 30 | 374 | 0 | 374 | 23 |
| 40 | 351 | 0 | 351 | 26 |
| 50 | 325 | 318 | 166 | 7 |
Example data
Based on the values in the table, let’s manually estimate the survival
function, density function, and hazard function together with their
standard errors.
- When r = 0
- When r = 1
= 14, = 0, = 432, = 432
- For r > 2, I will calculate them with R.
# input data
tau_interval <- 10
r <- 1:6
i <- seq(0, 50, 10)
Nr <- c(432, 418, 397, 374, 351, 325)
Zr <- c(0, 0, 0, 0, 0, 318)
Rr <- c(432, 418, 397, 374, 351, 166)
Er <- c(14, 21, 23, 23, 26, 7)
# by definition
qr <- Er / Rr
pr <- 1 - qr
hr <- Er / ((Rr - Er / 2) * tau_interval)
Sr <- c()
Sr[1] <- pr[1]
for (i in 2:6) {
Sr[i] <- Sr[i - 1] * pr[i]
}
fr <- c()
fr[1] <- 1 - Sr[1]
for (i in 2:6) {
fr[i] <- (Sr[i - 1] - Sr[i]) / tau_interval
}
# find se
se_sr <- c()
inner_sum <- qr[1] / (pr[1] * Rr[1])
se_sr[1] <- sqrt(Sr[1]^2 * inner_sum)
for (i in 2:6) {
inner_sum <- inner_sum + qr[i] / (pr[i] * Rr[i])
se_sr[i] <- sqrt(Sr[i]^2 * inner_sum)
}
se_fr <- c()
inner_sum <- qr[1] / (pr[1] * Rr[1])
se_fr[1] <- sqrt(((qr[1] * Sr[1] / tau_interval)^2) * (inner_sum + pr[1] / (qr[1] * Rr[1])))
# se_fr[1] = sqrt(((qr[1] * Sr[1]/ tau_interval)^2) * (1 + pr[1]/(qr[1] * Rr[1])))
for (i in 2:6) {
inner_sum <- inner_sum + qr[i] / (pr[i] * Rr[i])
se_fr[i] <- sqrt((qr[i] * Sr[i] / tau_interval)^2 * (inner_sum + pr[i] / (qr[i] * Rr[i])))
}
se_hr <- c()
for (i in 1:6) {
se_hr[i] <- sqrt((hr[i]^2 / (qr[i] * Rr[i])) * (1 - (hr[i] * tau_interval) / 2)^2)
}
df1 <- tibble::tibble(r, i, Nr, Zr, Rr, Er, qr, pr)
df2 <- tibble::tibble(Sr, se_sr, fr, se_fr, hr, se_hr)
| r | i | Nr | Zr | Rr | Er | qr | pr |
|---|---|---|---|---|---|---|---|
| 1 | 6 | 432 | 0 | 432 | 14 | 0.032 | 0.968 |
| 2 | 6 | 418 | 0 | 418 | 21 | 0.050 | 0.950 |
| 3 | 6 | 397 | 0 | 397 | 23 | 0.058 | 0.942 |
| 4 | 6 | 374 | 0 | 374 | 23 | 0.061 | 0.939 |
| 5 | 6 | 351 | 0 | 351 | 26 | 0.074 | 0.926 |
| 6 | 6 | 325 | 318 | 166 | 7 | 0.042 | 0.958 |
Example data
| Sr | se_sr | fr | se_fr | hr | se_hr |
|---|---|---|---|---|---|
| 0.968 | 0.009 | 0.032 | 0.001 | 0.003 | 0.001 |
| 0.919 | 0.013 | 0.005 | 0.001 | 0.005 | 0.001 |
| 0.866 | 0.016 | 0.005 | 0.001 | 0.006 | 0.001 |
| 0.812 | 0.019 | 0.005 | 0.001 | 0.006 | 0.001 |
| 0.752 | 0.021 | 0.006 | 0.001 | 0.008 | 0.001 |
| 0.721 | 0.023 | 0.003 | 0.001 | 0.004 | 0.002 |
Derived functions and corresponding standard error
The Kaplan-Meier (Product-Limit) Method
One helpful YouTube video to cover this method is from “zedstatistics”
here.
Its estimation is similar to life-table method above:
I really enjoy the fact that YouTube video step by step deducts the
stand error
.
Please check it out if interested!
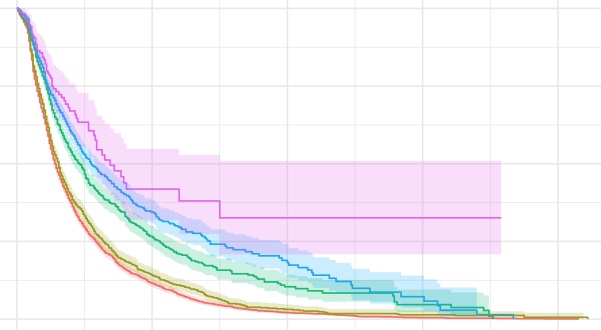
If you plot out the survival function versus the time, then you can get
the survival curve, which is often used in the observational data
analysis.
A simple exercise
The example data is from the survival analysis course at SU.
- To estimate the survival curves (Kaplan-Meier method), we need to use
survivallibrary in R.
library(survival)
library(tidyverse)
- Create a survival object with
Surv()function.
# read data
df <- readxl::read_excel("nonparametric_estimation/data/example_data.xlsx")
# create a survival object
birth_time <- Surv(df$ExpMonths, df$FirstBirth)
- Kaplan-Meier estimation for the different levels of birth cohort
km_BirCoh <- survfit(birth_time ~ as.factor(BirCoh), data = df)
- Plot the curve
plot(km_BirCoh, main = "Survival curve: Kaplan-Meier estimates")

- or we can have a better looking curve by using
survfit2from
ggsurvfitpackage.
library(ggsurvfit)
survfit2(birth_time ~ as.factor(BirCoh), data = df) %>%
ggsurvfit() +
labs(
x = "Months",
y = "Survival probability",
title = "Survival curve: Kaplan-Meier estimates",
color = "Birth cohort"
) +
theme(plot.title = element_text(hjust = 0.5)) +
add_confidence_interval()

Please note: since the confidence intervals are symmetric, when the
estimated survivor function is close to 0 or 1, they may sit outside the
(0,1) range. One solution is to manually enclose interval with 0 or 1.
This is also mentioned in the video above.
- The cumulative hazard function H(t) is estimated by
If we plot H(t) versus the time t, finding the slope is constant, then
it indicates that the hazard rate is constant over time.
Life-table VS Kaplan-Meier
- Life-table uses aggregated data, with arbitrary choice of interval.
This may lead to loss of information; Kaplan-Meier does not demand
grouping of data. It records every spot when event of interest
happens. Therefore we can get more information from Kaplan-Meier. - However, one shortcoming of Kaplan-Meier is that if one prints out
the estimations, it could be a very long table.
End thoughts
The derivation of life-table and Kaplan-Meier methods to estimate the
survival functions are intuitive and straightforward. It does not involve
complicated algebraic calculations. I feel the “statistical spices”
comes from the confidence interval and standard error estimation, which
are based on the theory of sampling distribution of the mean.
So now we know how to estimate the functions. In the next post, we will look at the comparison of two survival functions.